Welcome Home to SeNA Research
Subscribe for Updates & Offers
Selenium Nucleic Acids (SeNA) Technology
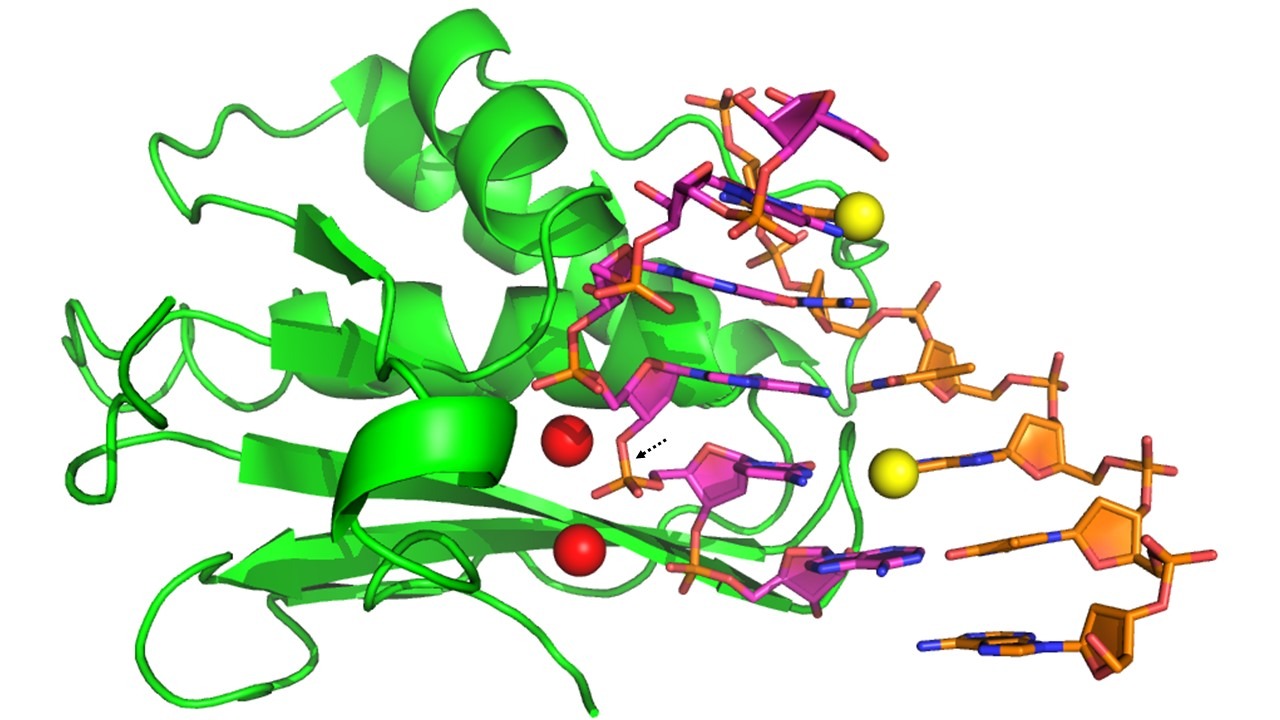
Figure 1
Our novel technologies for X-ray and neutron crystal structure determination and function studies of nucleic acids and protein-nucleic acid complexes have been discovered and developed by us. These exciting, convenient, and time-and-effort-saving methodologies using selenium-derivatized (or modified) nucleic acids (SeNA) can facilitate molecular packing and crystallization (such as fast crystal growth, broad buffer conditions, large crystals, and high-diffraction quality), and can allow phase and structure determination with high resolution (Figure 1-3). Selenium derivatization of nucleic acids has been proven superior to conventional methods as evidenced by:
• Faster crystal growth
• broader crystallization conditions
• Higher diffraction-quality crystals
• Larger crystal sizes
• More efficient phase determination
• Higher diffraction resolution
• Greater molecular stability during X-ray or neutron radiation and diffraction data collection
• Fine atomic labeling tools for cryo-EM, molecular and cellular imaging labeling, and mass
spectrometry research with selenium isotopes.
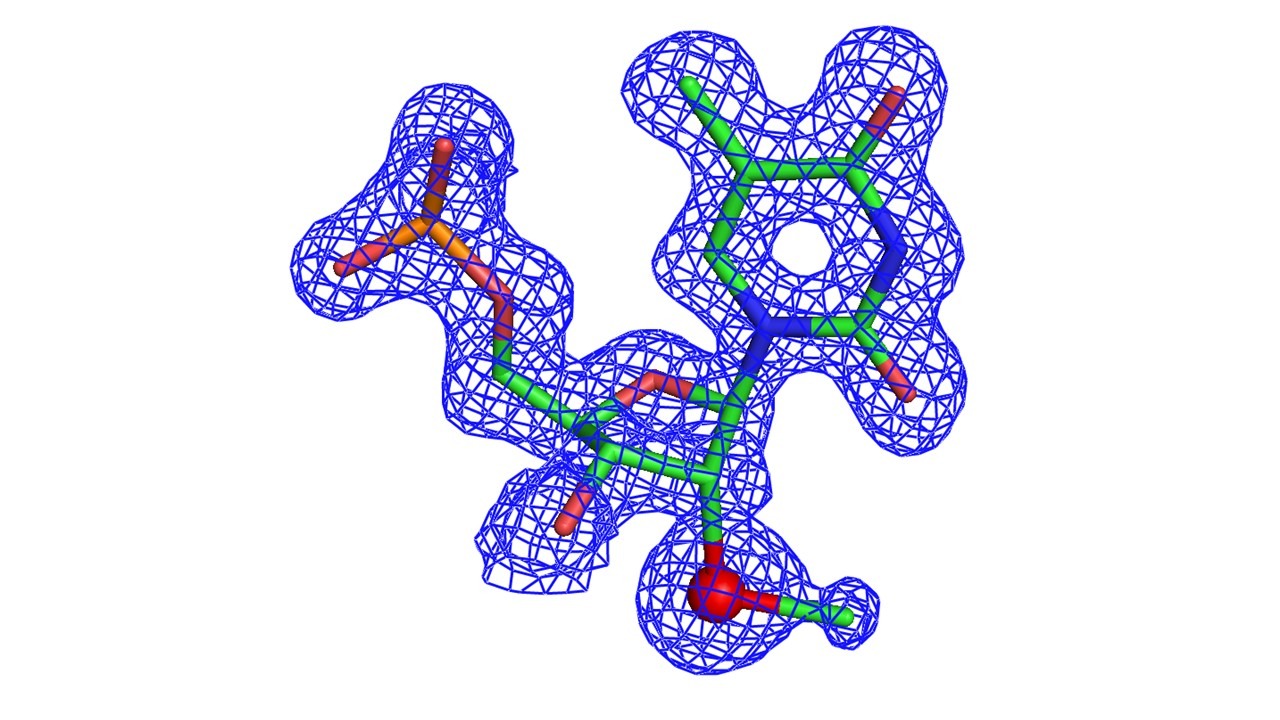
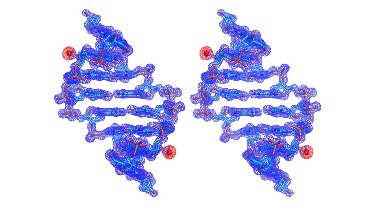
Figure 2 Figure 3
SeNA Research is established for its mission in developing and marketing products and services on the basis of our innovative and patented selenium technologies. These technologies include the atomic derivatization and modification of nucleic acids by replacing oxygen atom with selenium. We offer cutting-edge technologies to study X-ray crystal structures and functions of nucleic acids and their protein complexes (Figure 1).
Determining three-dimensional structures of nucleic acid molecules and their protein complexes is invaluable for understanding biological systems at the molecular level. Three-dimensional understanding is important because it dramatically facilitates new drug development and more efficient patient care. X-ray and neutron crystallography methods are among the most powerful tools for three-dimensional structure determination. Through 22 years of research, our team guided by Dr. Zhen Huang has successfully developed the novel selenium technologies to take advantage of the power of X-ray and neutron crystallography. These technologies involve derivatization strategies that replace oxygen atoms with selenium in nucleic acids. Replacing oxygen atom with selenium has several advantages over conventional methods of using bromine or iodine. One such advantage is that selenium can be selectively introduced to a variety of positions via oxygen replacement, rather than single-position capability for bromine and iodine. Proper selenium positioning can avoid disruption of structure and function caused by the modifications.
SeNA Facilitates Crystallization, Phase Determination & High Resolution
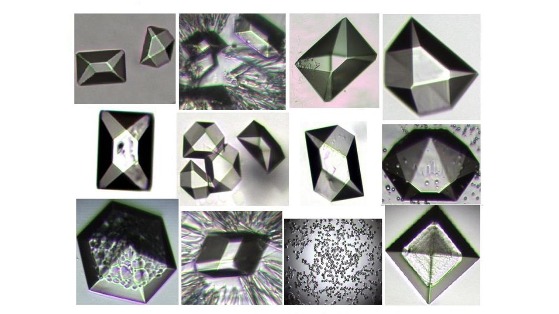
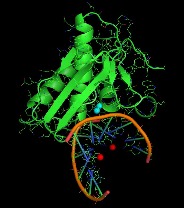
Figure 4: Enhanced Crystallization Figure 5: Phase Determination
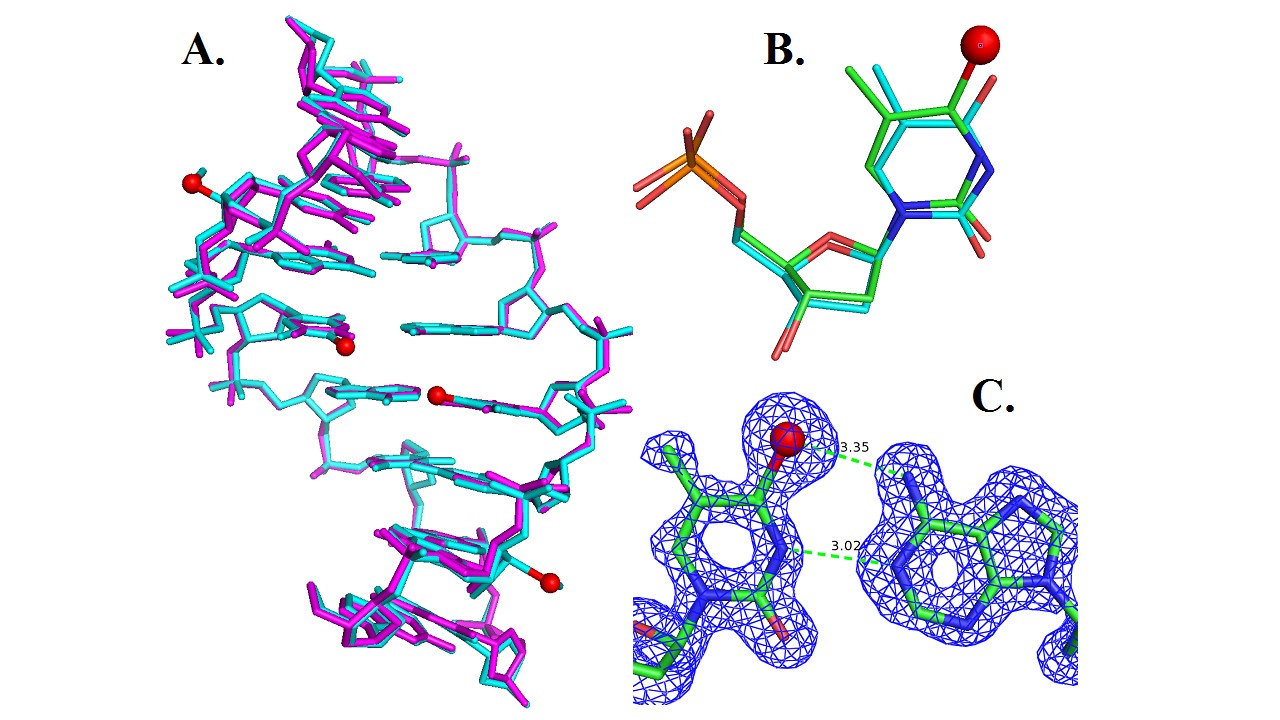
Figure 6: High-resolution Diffraction
3-Dimensional Structure determination of nucleic acids and protein-nucleic acid complexes (Figure 4-6) provides insight into how diseases initiate, manipulate and evolve at the molecular level. Understanding molecular structure 3-dimensionally facilitates new drug discovery and ultimately better patient care. X-ray and neutron crystallography methods are the most powerful and most accepted tool for 3-dimensional structure determination at atomic level. Incorporating selenium into RNA or DNA allows facilitation of
phase determination.Our innovative technologies have been established for discovering Novel Drugs targeting nucleic acids (such as RNA) and protein-nucleic acid complexes.
More Information on SeNA Technologies
Determination of the three-dimensional structures of RNA molecules, RNA-protein and DNA-protein complexes with high resolution is invaluable for gaining understanding of biological systems at the molecular level. X-ray crystallography is the most direct and powerful tool for structure determination of these macromolecules. Besides the difficulties related to crystallization, however, heavy atom derivatization for phase determination, a long-standing problem in nucleic acid X-ray crystallography, has largely slowed down structural determination of new structures and folds. The conventional approaches for DNA and RNA derivatization, such as heavy-atom soaking and co-crystallization, have proved to be much more difficult for nucleic acids than for proteins, probably because nucleic acids often lack specific binding sites for metal ions. In addition, the radiation stability and structure perturbation have reduced the usefulness of the halogen derivatization (such as Br and I).
Since 1998, Dr. Zhen Huang’s research group has pioneered selenium derivation of nucleic acids for X-ray crystallographic studies (Ref. 1). So far, we have successfully developed various chemical and enzymatic strategies to atom-specifically substitute oxygen with selenium and covalently incorporate Se-functionality into nucleic acids at a variety of positions, including 5’, 2’, nucleobase and phosphate non-bridging positions (Ref. 1-12). Through collaboration among Huang, Egli and co-workers, we have determined for the first time structure of a nucleic acid covalently-derivatized with selenium via MAD phasing (Ref. 3). This atom-specific derivatization with selenium leads to synthesis of selenium-derivatized nucleic acids (SeNA), which have great potential in structural and functional studies of nucleic acids. Unlike conventional halogen derivatization (Br or I), where halogens are primarily placed on the 5-position of deoxyuridine (a mimic of thymidine), selenium can be selectively introduced to a variety of positions via oxygen replacement (e.g., 2'-, 3'-, 5'-ribose oxygen, furan ring oxygen, non-bridging phosphate oxygen, or oxygen on nucleobases; Ref. 1-12). Choice of positioning can avoid disruption of structure and function caused by modification. We have found that the 2’-Se-derivatized DNA structure reveals that the 2’-Se-furanose displays the 3’-endo sugar pucker (Fig. 1 & 2), which is consistent with the sugar pucker of A-form DNAs and RNAs, and the 2’-methylseleno group is placed in the minor groove of the duplex.
We have also successfully crystallized the 4-Se-T-containing DNA (5’-G-dUSe-G-SeT-A-C-A-C-3’, self-complementary), where the dUSe (2’-Se-dU) was used to facilitate the crystal growth (Ref. 11). The determined crystal structure (1.50 Å resolution) of the Se-DNA is superimposable over the native structure in the same tetragonal space group, indicating that the O replacement by Se does not cause the significant structure perturbation (Fig. 3). The large Se atom is accommodated by a slight shift, revealing the flexibility of the DNA duplex structure. Furthermore, the atomic distance (3.02 Å) between the thymine N3 and the adenine N1 indicates a hydrogen bond formation. In addition, considering that the Se atomic radius is 0.43 Å larger than that of O, and that the distance between T4 exo-O4 and A5 exo-N6 in the native structure is 2.87 Å, the distance (3.35 Å) between the exo-Se4 and exo-N6 indicates a Se-mediated hydrogen bond formation.
In addition, we found that the local backbone torsion angles and solvent hydration patterns were altered in the DNA structure with the Br derivatization (Ref. 10). Furthermore, while the native and Br-derivatized DNAs needed over several weeks to form reasonable-size crystals, we observed that the Se-derivatized DNAs grew crystals overnight with high diffraction quality (Ref. 9 and 10), suggesting that the Se-derivatization facilitated the crystal formation. Moreover, the Se-derivatized DNA sequences crystallized under a broader range of buffer conditions, and generally had a faster crystal growth rate. We have observed that the Se-DNA crystal formation in a few days or even overnight, while the crystals of the corresponding native DNAs need two to three months to grow. Our experimental results indicate that the selenium derivatization of DNAs may largely facilitate the determination of nucleic acid X-ray crystal structures in phasing and high-quality crystal growth. Our results suggest that the Se derivatization will be a better alternative to the conventional Br derivatization. This Se derivatization strategy via the atom-specific substitution will significantly facilitate X-ray crystal structure studies of nucleic acids and their protein complexes.
References:
52. Bei Hu, Yitao Wang, Shichao Sun, Weizhu Yan, Chong Zhang, Danyan Luo, Huiling Deng, Lillian R. Hu, Zhen Huang*, “Synthesis of Selenium-triphosphates (dNTPaSe) for morespecific DNA polymerization”, Angew. Chem. Int. Ed., 2019, 58, 7835-7839.
51. Yiqing Chen, Hehua Liu, Chun Yang, Yanqing Gao, Xiang Yu, Xi Chen, Ruixue Cui, Lina Zheng, Suhua Li, Xuhang Li, Jinbiao Ma, Zhen Huang*, Jixi Li* and Jianhua Gan*, “Structure of the error-prone DNA ligase of African swine fever virus identifies critical active site residues”, Nature Communications, 2019, 10, 387.
50. Xinrui Zhoua, Weizhu Yana, Chong Zhanga, Zhaoyi Yanga, Peter Neubauerb*, Igor A. Mikhailopulo*, Zhen Huang*, Biocatalytic synthesis of seleno-, thio- and chloro-nucleobase modified nucleosides by thermostable nucleoside phosphorylases, Catalysis Communications, 2019, 121, 32-37.
49. Venu Gopal Vandavasi, Matthew P. Blakeley, David A. Keen, Lillian R. Hu, Zhen Huang* and Andrey Kovalevsky*, “Temperature-induced replacement of phosphate proton with metal ion captured in neutron structures of A-DNA”, Structure, 2018, 26, 1-6.
48. Hehua Liu, Xiang Yu, Yiqing Chen, Jing Zhang, Baixing Wu, Lina Zheng, Phensinee Haruehanroengra, Rui Wang, Suhua Li, Jinzhong Lin, Jixi Li, Jia Sheng, Zhen Huang*, Jinbiao Ma*, and Jianhua Gan*, “Crystal structure of an RNA-cleaving DNAzyme”, Nature Communications, 2017, 8, 2006.
47. Yiqing Yiqing Chen, Jing Zhang, Hehua Liu, Yanqing Gao, Xuhang Li, Lina Zheng, Ruixue Cui, Qingqing Yao, Liang Rong, Jixi Li, Zhen Huang*, Jinbiao Ma* and Jianhua Gan*, “Unique 50-P recognition and basis for dG:dGTP misincorporation of ASFV DNA polymerase X”, PLoS Biology, 2017, 15, e1002599.
46. Wen Zhang, Jack W. Szostak, and Zhen Huang*, “Nucleic Acid Crystallization and X-ray Crystallography Facilitated by Single Selenium Atom”, 2016, Frontiers of Chemical Science and Engineering, 10, 196-202.
45. Liqin Zhang, Zunyi Yang, Kwame Sefah, Kevin M. Bradley, Shuichi Hoshika, Myong-Jung Kim, Hyo-Joong Kim, Guizhi Zhu, Sena Cansiz, I-Ting Teng, Carole Champanhac, Christopher McLendon, Chen Liu, Wen Zhang, Dietlind Gerloff3, Zhen Huang*, Weihong Tan* and Steven A. Benner*, “Crystal Structure, Evolution, and Function of Six-Nucleotide DNA. Exploring its Large Sequence Space”, 2015, Journal of American Chemical Society, 137, 6734-6737.
44. Huiyan Sun and Zhen Huang*, "Nucleic Acid Crystallography via Direct Selenium Derivatization: RNAs Modified with Se-Nucleobases" (invited and peer reviewed), 2015, Nucleic Acid Crystallography: Methods and Protocols (Humana Press), Chapter 12, 193-204.
43. Rob Abdur, Oksana O. Gerlits, Jianhua Gan, Jiansheng Jiang, Jozef Salon, Andrey Y. Kovalevsky, Alexander Chumanevich, Irene T. Weber, Zhen Huang*, “Novel Complex MAD Phasing and RNase H Structural Insights by Selenium Oligonucleotides”, 2014, Acta Crystallographica Section D, D70, 354-361.
42. Manindar Kaur and Zhen Huang*, “Synthesis and optical behaviors of 6-seleno-deoxyguanosine”, Science China: Chemistry, 2014, 57, 314-321.
41. Jia Sheng, Jianhua Gan, Alexie Soars, Jozef Salon and Zhen Huang*, “Structural Insights of Non-canonical U•U Pair and Hoogsteen Interaction Probed with Se Atom”, 2013, Nucleic Acids Research, 41, 10476-10487.
40. Jia Sheng, Jianhua Gan and Zhen Huang*, “Structure-Based DNA-Targeting Strategies with Small Molecule Ligands for Drug Discovery”, Medicinal Research Reviews, 2013, 33, 1119-1173.
39. Huiyan Sun, Sibo Jiang, Julianne Caton-Williams, Hehua Liu, and Zhen Huang*, “2-Selenouridine Triphosphate Synthesis and Se-RNA Transcription”, RNA, 2013, 19, 1309-1314.
38. Jozef Salon, Jianhua Gan, Rob Abdur, Hehua Liu and Zhen Huang*, “Synthesis of 6-Se-Guanosine RNAs for Structural Study”, Organic Letter, 2013, 15, 3934-3937.
37. Wen Zhang, Abdalla E. Hassan, and Zhen Huang*, “Synthesis of Novel Di-Se-containing Thymidine and Se-DNAs for Structure and Function Studies”, Science China: Chemistry, 2013, 56, 273-278.
36. Manindar Kaur, Abdur Rob, Julianne Caton-Williams, and Zhen Huang*, “Biochemistry of the nucleic acids functionalized with sulfur, selenium and tellurium: Roles of the single-atom substitution” (invited and peer reviewed), Biochalcogen Chemistry: The Biological Chemistry of Sulfur, Selenium, and Tellurium; Bayse, C., et al.; ACS Symposium Series; American Chemical Society: Washington, DC, 2013; ACS Books Publisher, 2013, 89-126.
35. Julianne Caton-Williams, Rudiona Hoxhaj, Bilal Fiaz and Zhen Huang*, “Use of a Novel 5′-Regioselective Phosphitylating Reagent for One-Pot Synthesis of Nucleoside 5′-Triphosphates from Unprotected Nucleosides”, Current Protocols in Nucleic Acid Chemistry (invited and peer reviewed), John Wiley & Sons, Inc., 2013, Chapter 1:Unit1.30.
34. Sibo Jiang, Huiyan Sun and Zhen Huang*, “Selenium Atom-specific Mutagenesis (SAM) for DNA Nanostructure Design and Application” (invited and peer reviewed), DNA Nanotechnology: From Structure to Function, Springer, 2013, 29-56.
33. Huiyan Sun and Zhen Huang*, “Atom-specific Mutagenesis of RNAs for Structure, Function and Therapeutics Studies” (invited and peer reviewed), RNA Nanotechnology and Therapeutics, John Wiley & Sons, Inc., 2013, 213-234.
32. Jia Sheng, Wen Zhang, Abdalla E. A. Hassan, Jianhua Gan, Alexei Soares, Song Geng, Yi Ren, Zhen Huang*, "Hydrogen Bond Formation between the Naturally Modified Nucleobase and Phosphate Backbone", Nucleic Acids Research, 2012, 40, 8111-8118.
31. Huiyan Sun, Jia Sheng, Abdalla E. A. Hassan, Sibo Jiang, Jianhua Gan and Zhen Huang*, “Novel RNA Base Pair with Higher Specificity using Single Selenium Atom”, Nucleic Acids Res., 2012, 40, 5171-5179.
30. Julianne Caton-Williams, Bilal Fiaz, Rudiona Hoxhaj, Matthew Smith, and Zhen Huang*, “Convenient Synthesis of Nucleoside 5’-(α-P-thio)triphosphates and phosphorothioate nucleic acids (DNA and RNA)”, Science China: Chemistry, 2012, 55, 80-89.
29. Wen Zhang, Jia Sheng, Abdalla E. Hassan, and Zhen Huang*, "Synthesis of Novel 2’-Deoxy-5-(Methylselenyl)Cytidine and Se-DNAs for Structure and Function Studies", Chemistry-An Asian Journal, 2012, 7, 476-479.
28. Jia Sheng and Zhen Huang*, "Selenium Derivatization of Nucleic Acids for X-ray Crystal Structure and Function Studies", (invited and peer reviewed), in “Chemistry and Biology of Artificial Nucleic Acids” Ed. by Martin Egli and Piet Herdewijn, John Wiley & Sons, Inc., 2012, 329-361, Zurich, Switzerland.
27. Lina Lin and Zhen Huang*, "Se-derivatized RNAs for X-ray Crystallography" (invited and peer reviewed), Preparation of RNA for Biochemical and Biophysical Analysis: Methods and Protocols (Humana Press), 2012, 941:213-225.
26. Julianne Caton-Williams, Matthew Smith, Nicolas Carrasco and Zhen Huang*, "Protection-free One-pot Synthesis of 2’-Deoxynucleoside 5’-Triphosphates for DNA Polymerization", Organic Letter, 2011, 13, 4156-4159.
25. Jia Sheng, Abdalla E. A. Hassan, Wen Zhang, Jianfeng Zhou, Bingqian Xu, Alexei S. Soares and Zhen Huang*, "Synthesis, Structure and Imaging of Oligodeoxyribonucleotides with Tellurium-nucleobase Derivatization", Nucleic Acids Research, 2011, 39, 3962-3971.
24. Wen Zhang and Zhen Huang*, "Synthesis of the 5’-Se-Thymidine Phosphoramidite and Convenient Labeling of DNA Oligonucleotide", Organic Letter, 2011, 13, 2000-2003.
23. Julianne Caton-Williams, Lina Lin, Mathew Smith, and Zhen Huang*, "Convenient Synthesis of Nucleoside 5’-Triphosphates for RNA Transcription", Chemical Communications, 2011, 47, 8142-8144.
22. Lina Lin, Julianne Caton-Williams, Manindar Kaur, Andres M. Patino, Jia Sheng, Jaya Punetha, and Zhen Huang*, "Facile Synthesis of Nucleoside 5’-(alpha-P-seleno)-triphosphates and Phosphoroselenoate RNA Transcription", RNA, 2011, 1932-1938.
21. Lina Lin, Jia Sheng, and Zhen Huang*, "Nucleic Acid X-ray Crystallography via Direct Selenium Derivatization", Chemical Society Reviews (invited and peer reviewed), 2011, 40, 4591-4602.
20. Jozef Salon, Bo Zhang, Zhen Huang,* "Mild Detritylation of Nucleic Acid Hydroxyl Groups by Warming-up", Nucleosides, Nucleotides and Nucleic Acids, 2011, 30, 271-279.
19. Wen Zhang, Jia Sheng, and Zhen Huang*, "Structures and Functions of Nucleic Acids Modified with S, Se and Te and Complexed with Small Molecules" (invited and peer reviewed), Medicinal Chemistry of Nucleic Acids (John Wiley & Sons, Inc.), 2011, 101-141.
18. Jianhua Gan, Jia Sheng, and Zhen Huang*, “Chemical and Structural Biology of Nucleic Acids and Their Protein Complexes for Novel Drug Discovery”, (invited and peer reviewed), Science China: Chemistry, 2011, 54, 3-23. ##
17. Abdalla E. A. Hassan, Jia Sheng, Wen Zhang, and Zhen Huang*, "High Fidelity of Base Paring by 2-Selenothymidine in DNA", Journal of American Chemical Society, 2010, 132, 2120–2121.
16. Jia Sheng and Zhen Huang*, "Selenium Derivatization of Nucleic Acids for X-ray Crystal Structure and Function Studies", (invited and peer reviewed), Chemistry and Biodiversity (John Wiley & Sons, Inc.), 2010, 7, 753-785.
15. Jozef Salon, Jia Sheng, Jianhuan Gan, Zhen Huang*, "Synthesis and crystal structure of 2’-Se-modified guanosine containing DNA", 2010, J. Org. Chem., 75, 637–641.
14. Jia Sheng, Jozef Salon, Jianhua Gan, Zhen Huang*, "Synthesis and Crystal Structure Study of 2’-Se-Adenosine-Derivatized DNA", Science China: Chemistry, 2010, 53, 78–85.
13. Abdalla E. A. Hassan, Jia Sheng, Jiansheng Jiang, Wen Zhang, and Zhen Huang*, “Synthesis and Crystallographic Analysis of 5-Se-Thymidine DNAs”, Organic Letter, 2009, 11, 2503-2506.
12. Julianne Caton-Williams and Zhen Huang*, "Synthesis of Colored 4-Selenothymidine Triphosphate and its DNA Polymerase Incorporation into DNAs for Visualization, Structure and Function Studies", Angew. Chem. Int. Ed., 2008, 47, 1723-1725.
11. Jozef Salon, Jia Sheng, Jiansheng Jiang, Guexiong Chen, Julianne Caton-Williams, Zhen Huang*, “Oxygen Replacement with Selenium at the Thymidine 4-position for the Se-Base-Paring and Crystal Structure Studies”, Journal of American Chemical Society, 2007, 129, 4862-4863.
10. Jiansheng Jiang, Jia Sheng, Nicolas Carrasco, and Zhen Huang*, “Selenium Derivatization of Nucleic Acids for Crystallography”, Nucleic Acids Research, 2007, 35, 477-485.
9. Jia Sheng, Jiansheng Jiang, Jozef Salon, and Zhen Huang*, “Synthesis of a 2’-Se-Thymidine Phosphoramidite and Its Incorporation into Oligonucleotides for Crystal Structure Study”, Organic Letters, 2007, 9, 749-752.
8. Nicolas Carrasco, Julianne Canton-Williams, Gary Brandt, Siming Wang, and Zhen Huang*, “Efficient Enzymatic Synthesis of Phosphoroselenoate RNA Using Adenosine 5’-(a-P-seleno)triphosphate”, Angewandte Chemie Int. Ed., 2006, 45, 94-97.
7. Jozef Salon, Guexiong Chen, Yoani Portilla, Markus W. Germann, and Zhen Huang*, “Synthesis of a Novel 2’-Se-uridine Phosphoramidite and Its Incorporation into Oligonucleotides for Structure Study”, Organic Letter, 2005, 7, 5645-5648.
6. Yuri Buzin, Nicolas Carrasco, and Zhen Huang*, “Synthesis of Selenium-Derivatized Cytidine and Oligonucleotides for X-ray Crystallography Using MAD”, Organic Letters, 2004, 6, 1099-1102.
5. Nicolas Carrasco, Yuri Buzin, Elizabeth Tyson, Elizer Halpert, and Zhen Huang*, “Selenium derivatization and crystallization of DNA and RNA oligonucleotides for X-ray crystallography using multiple anomalous dispersion”, Nucleic Acids Research, 2004, 32, 1638-1646.
4. Nicolas Carrasco and Zhen Huang*, “Enzymatic Synthesis of Phosphoroselenoate DNA using Thymidine 5’-(a -P-Seleno)-triphosphate and DNA Polymerase for X-ray Crystallography using MAD”, Journal of American Chemical Society, 2004, 126, 448-449.
3. Marianna Teplova, Christopher J. Wilds, Quan Du, Nicolas Carrasco, Zhen Huang, and Martin Egli*, "Covalent Incorporation of Selenium into Oligonucleotides for X-ray Crystal Structure Determination via MAD: Proof of Principle", Biochimie, 2002, 84, 849-858.
2. Quan Du, Nicolas Carrasco, Marlanna Teplova, Christopher J. Wilds, Martin Egli, and Zhen Huang*, “Internal Derivatization of Oligonucleotides with Selenium for X-ray Crystallography Using MAD”, Journal of American Chemical Society., 2002, 124, 24-25.
1. Nicolas Carrasco, Dov Ginsburg, Quan Du, and Zhen Huang*, “Synthesis of Selenium-Derivatized Nucleosides and Oligonucleotides for X-ray Crystallography”, Nucleosides, Nucleotides, & Nucleic Acids, 2001, 20, 1723-1734.
Products & Services
Contact us for Selenium-, Sulfur- or Tellurium-modifiedNucleosides & Nucleotides:
Request a quote at Se_Nucleic_Acids@SeNAresearch.com
Services:
1. Custom synthesis of Se-, S- or Te-nucleic acids (DNA and RNA)
2. Custom Synthesis of Se-, S- or Te-nucleoside Triphosphates
3. Custom Synthesis of Se-, S- or Te-nucleoside Phosphoramidites
4. Custom Service on crystallization of nucleic acids and protein-nucleic acid complexes.
5. Custom service on crystal structure determination of nucleic acids and protein-nucleic acid complexes.
Products, Se-Phosphoramidites:
2'-Se-A-CE Phosphoramidite
2'-Se-C-CE Phosphoramidite
2'-Se-G-CE Phosphoramidite
2'-Se-T-CE Phosphoramidite
2'-Se-U-CE Phosphoramidite
2-Se-T-CE Phosphoramidite
4-Se-T-CE Phosphoramidite
5-Se-T-CE Phosphoramidite
2-Se-U-CE Phosphoramidite
4-Se-U-CE Phosphoramidite
6-Se-G-CE Phosphoramidite
HPLC-purified Sp & Rp isomers (mixed or individual one):
alpha-Se-dATP (dATPaSe)
alpha-Se-dCTP (dCTPaSe)
alpha-Se-dGTP (dGTPaSe)
alpha-Se-TTP (TTPaSe)
alpha-Se-ATP (ATPaSe)
alpha-Se-CTP (CTPaSe)
alpha-Se-GTP (GTPaSe)
alpha-Se-UTP (UTPaSe)
alpha-S-dATP (dATPaS)
alpha-S-dCTP (dCTPaS)
alpha-S-dGTP (dGTPaS)
alpha-S-TTP (TTPaS)
alpha-S-ATP (ATPaS)
alpha-S-CTP (CTPaS)
alpha-S-GTP (GTPaS)
alpha-S-UTP(UTPaS)
Items for Triphosphate Synthesis:
3H-1, 2-benzothiaselenol-3-one (BTSe; 3H-1, 2-benzothaselenol-3-one missing “i”)
2-chloro-4H-1,3,2-benzodioxaphosphorin-4-one
tris-(tributylammonium) pyrophosphate
tris-(tripropylammonium) pyrophosphate

